Mitochondrial AOX Supports Redox Balance of Photosynthetic Electron Transport, Primary Metabolite Balance, and Growth in Arabidopsis thaliana under High Light
Abstract
:1. Introduction
2. Results
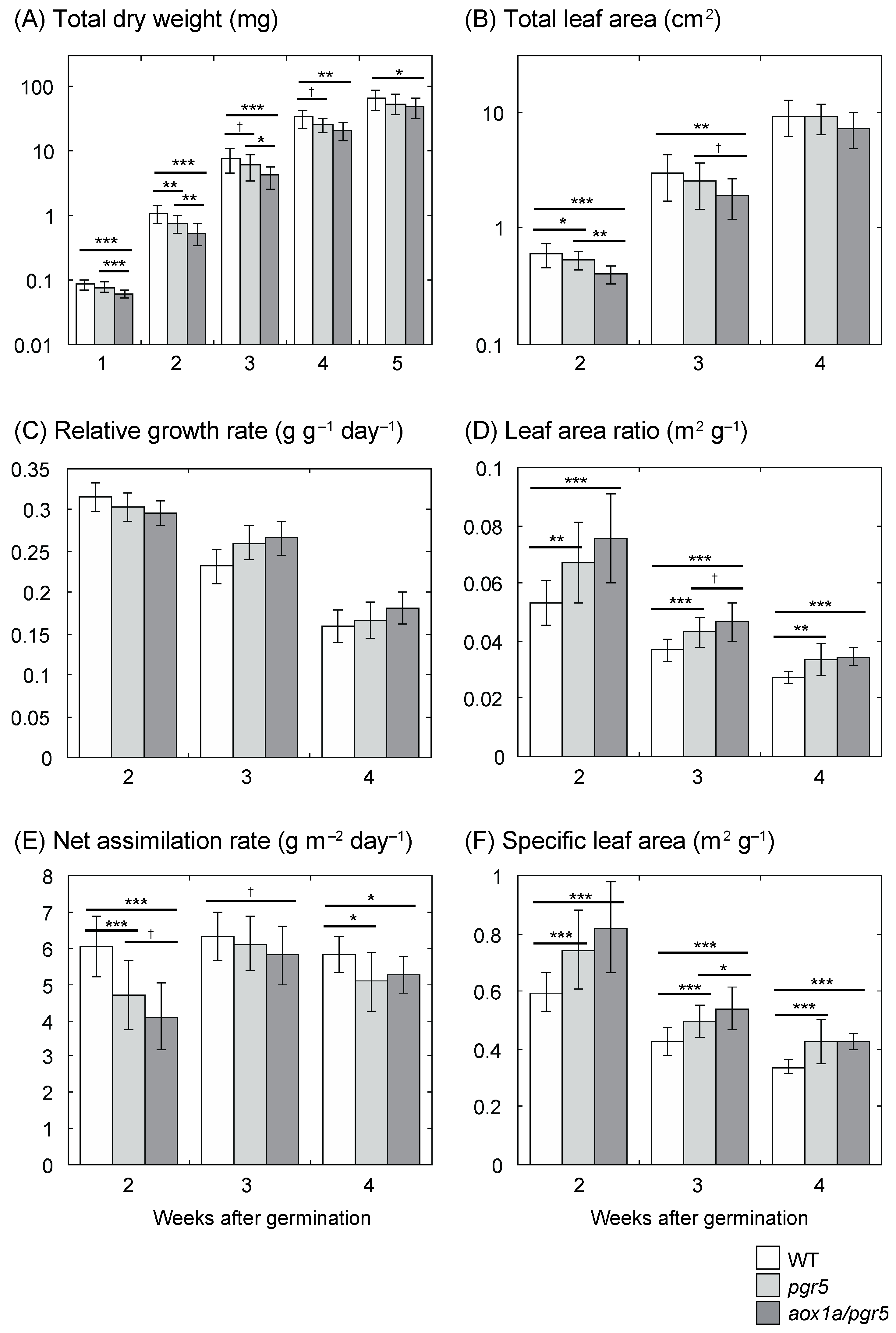
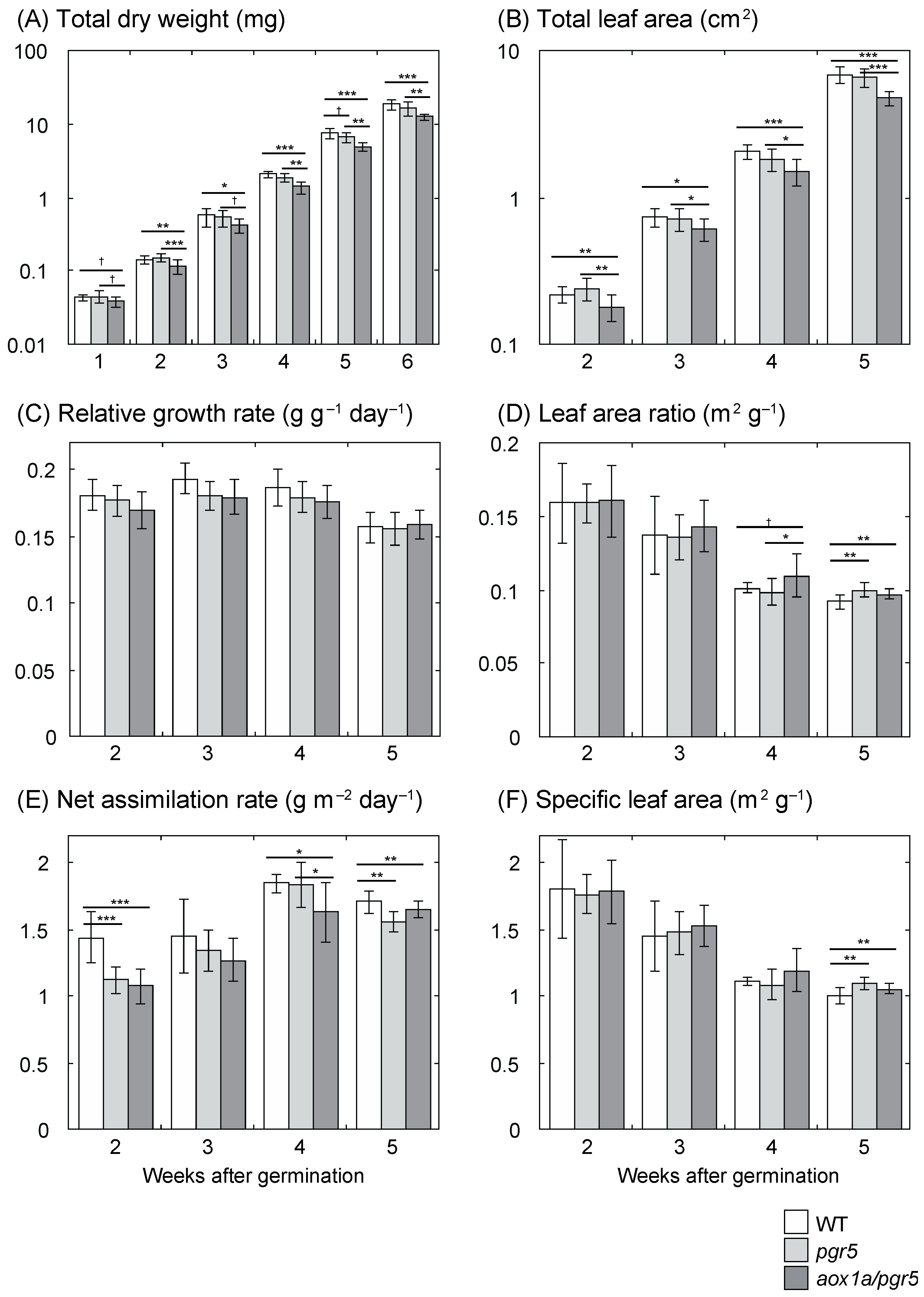
2.1. Low Net Assimilation Rate in aox1a/pgr5
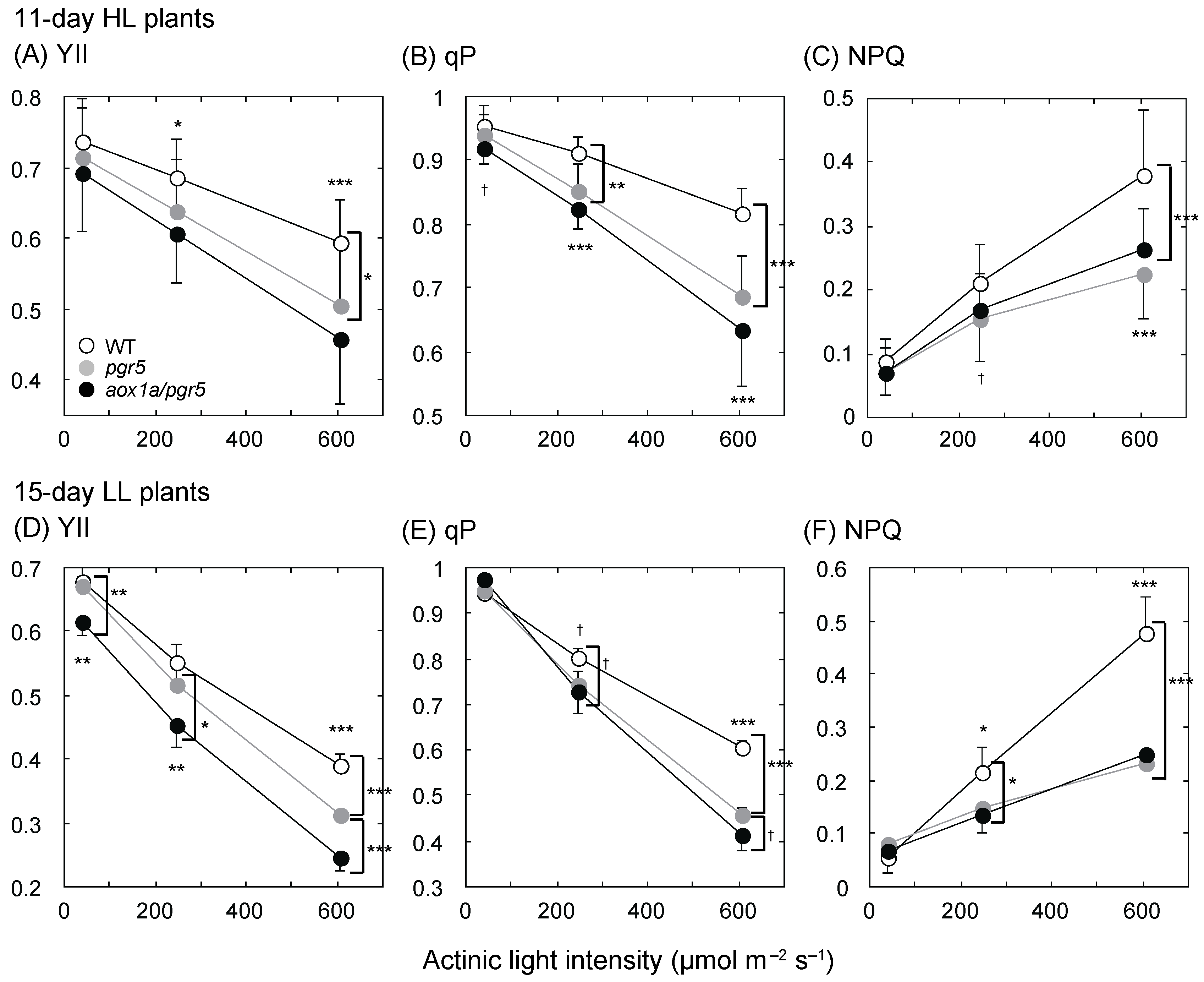
2.2. PSII Parameters Are Lower in aox1a/pgr5 under High Actinic Light Intensity
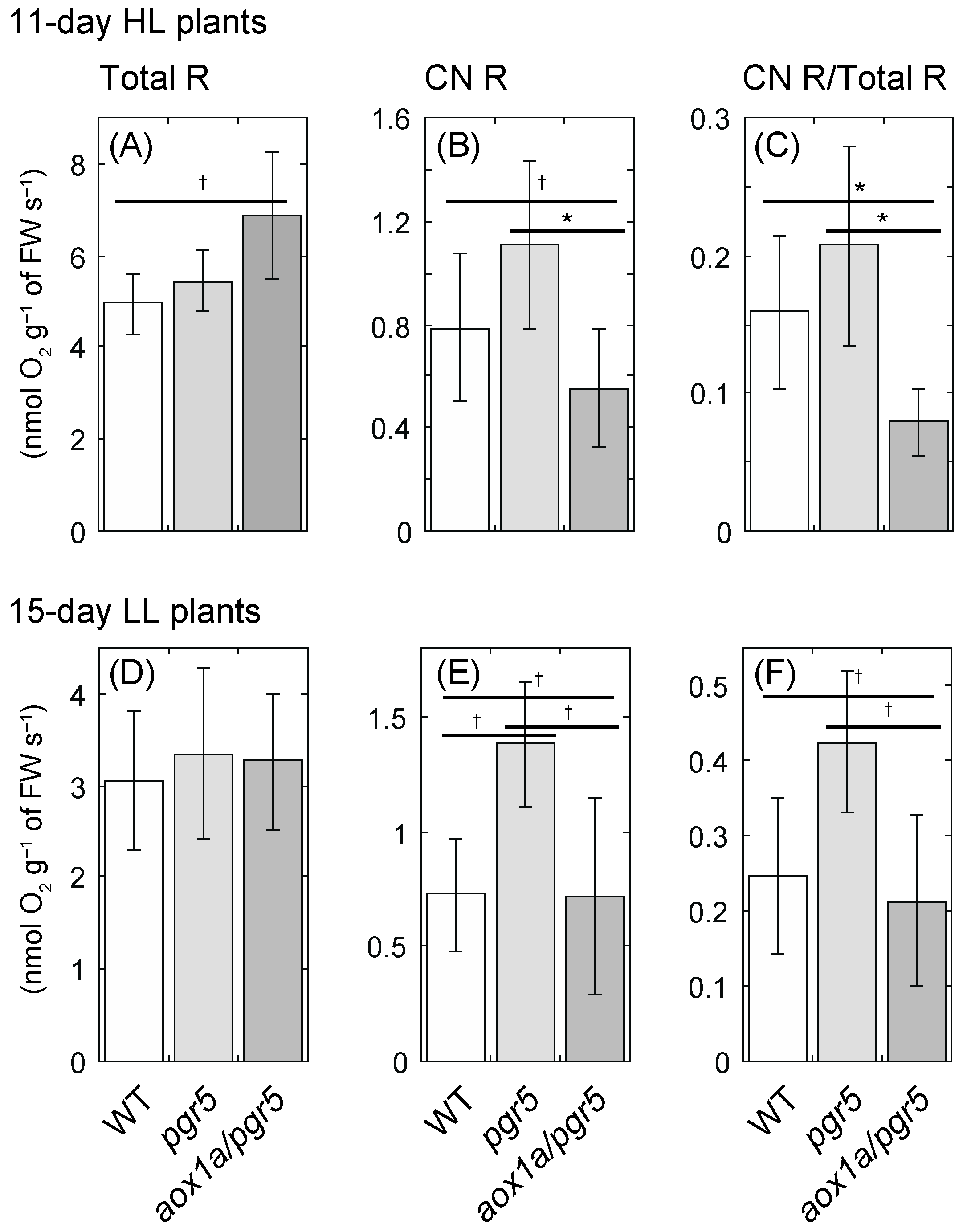
2.3. Cyanide-Resistant Respiratory O2 Uptake Rate Is Upregulated in pgr5 Mutant, Not in aox1a/pgr5
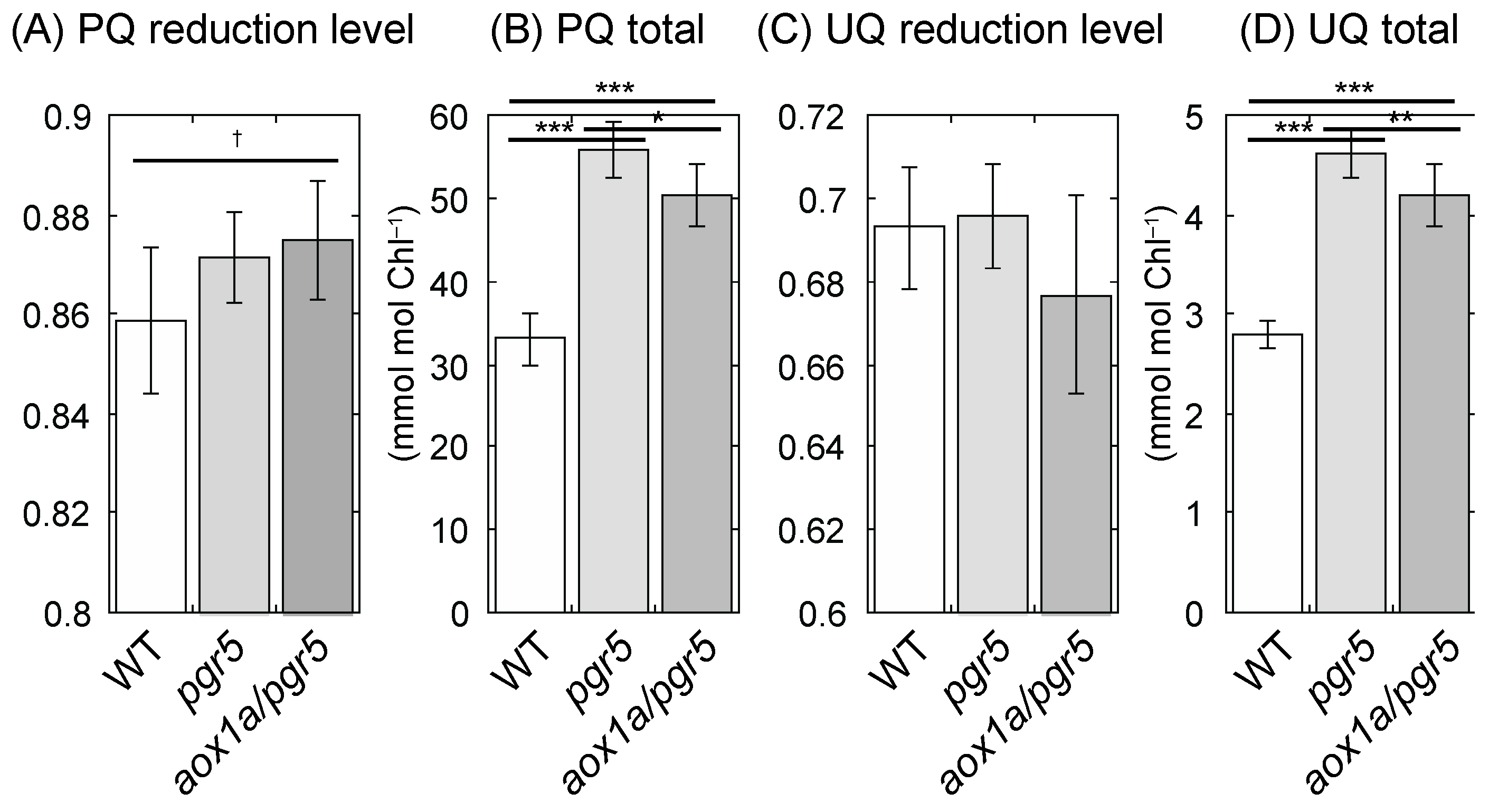
2.4. PQ Is Reduced in aox1a/pgr5 under HL
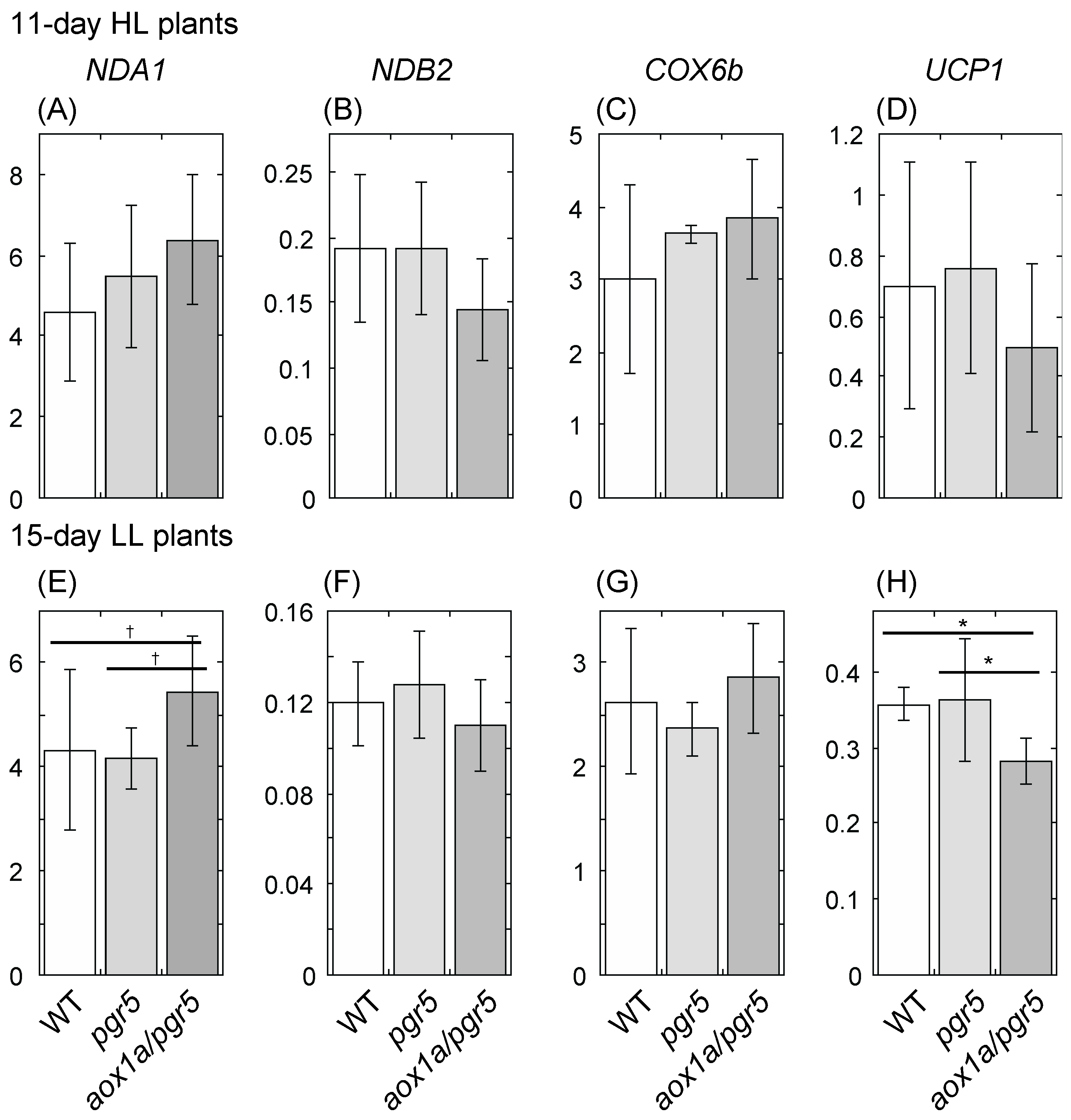
2.5. Gene Expression of the Respiratory Chain Is Partly Changed in aox1a/pgr5
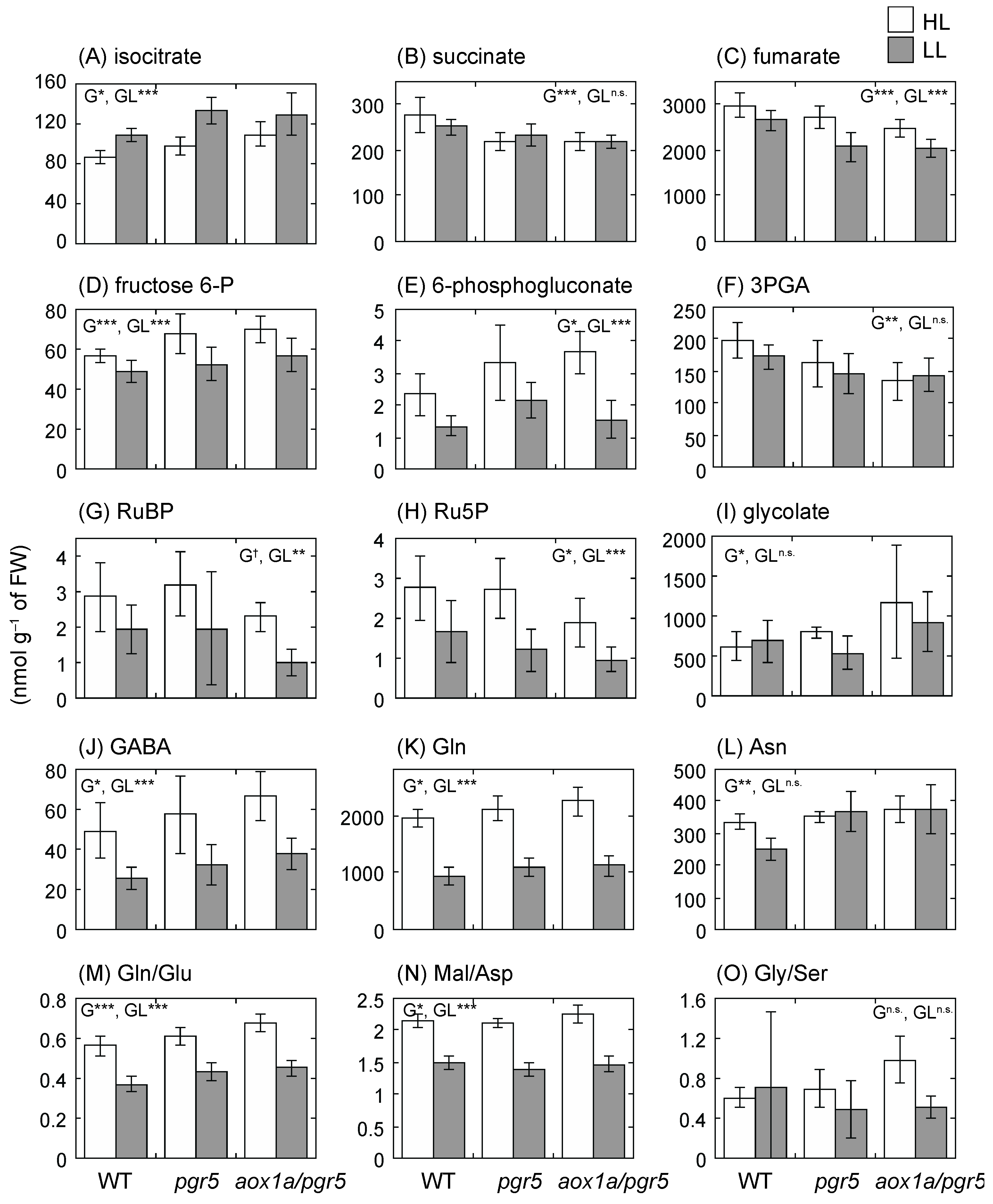
2.6. Primary Metabolite Levels Are Unbalanced in aox1a/pgr5
3. Discussion
3.1. Low Net Assimilation Rate in aox1a/pgr5 at the Early Growth Stage
3.2. Changes in Photosynthetic Electron Transport and Respiratory Chain in aox1a/pgr5
3.3. Changes in Primary Metabolite Balance in aox1a/pgr5
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Growth Analysis
4.3. Chlorophyll Fluorescence Measurements
4.4. Measurement of Respiratory O2 Uptake Rate
4.5. Quinone Determination
4.6. Real-Time PCR
4.7. Metabolome Analysis
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Raghavendra, A.S.; Padmasree, K. Beneficial interactions of mitochondrial metabolism with photosynthetic carbon assimilation. Trends Plant Sci. 2003, 8, 546–553. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, K.; Yoshida, K. Interaction between photosynthesis and respiration in illuminated leaves. Mitochondrion 2008, 8, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Araujo, W.L.; Nunes-Nesi, A.; Fernie, A.R. On the role of plant mitochondrial metabolism and its impact on photosynthesis in both optimal and sub-optimal growth conditions. Photosyn. Res. 2014, 119, 141–156. [Google Scholar] [CrossRef] [PubMed]
- Sunil, B.; Saini, D.; Ramesh, R.B.; Bapatla, R.B.; Aswani, V.; Raghavendra, A.S. Photorespiration is complemented by cyclic electron flow and the alternative oxidase pathway to optimize photosynthesis and protect against abiotic stress. Photosyn. Res. 2019, 139, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Del-Saz, N.F.; Ribas-Carbo, M.; McDonald, A.E.; Lambers, H.; Fernie, A.R.; Florez-Sarasa, I. An in vivo perspective of the role(s) of the alternative oxidase pathway. Trends Plant Sci. 2018, 23, 206–219. [Google Scholar] [CrossRef] [PubMed]
- Millar, A.H.; Whelan, J.; Soole, K.L.; Day, D.A. Organization and regulation of mitochondrial respiration in plants. Annu. Rev. Plant Biol. 2011, 62, 79–104. [Google Scholar] [CrossRef] [PubMed]
- Vanlerberghe, G.C. Alternative oxidase: A mitochondrial respiratory pathway to maintain metabolic and signaling homeostasis during abiotic and biotic stress in plants. Int. J. Mol. Sci. 2013, 14, 6805–6847. [Google Scholar] [CrossRef] [PubMed]
- Clifton, R.; Lister, R.; Parker, K.L.; Sappl, P.G.; Elhafez, D.; Millar, A.H.; Day, D.A.; Whelan, J. Stress-induced co-expression of alternative respiratory chain components in Arabidopsis thaliana. Plant Mol. Biol. 2005, 58, 193–212. [Google Scholar] [CrossRef]
- Wanniarachchi, V.R.; Dametto, L.; Sweetman, C.; Shavrukov, Y.; Day, D.A.; Jenkins, C.L.D.; Soole, K.L. Alternative respiratory pathway component genes (AOX and ND) in rice and barley and their response to stress. Int. J. Mol. Sci. 2018, 19, 915. [Google Scholar] [CrossRef]
- Watanabe, C.K.; Hachiya, T.; Terashima, I.; Noguchi, K. The lack of alternative oxidase at low temperature leads to a disruption of the balance in carbon and nitrogen metabolism, and to an up-regulation of anti-oxidant defence systems in Arabidopsis thaliana leaves. Plant Cell Environ. 2008, 31, 1190–1202. [Google Scholar] [CrossRef]
- Watanabe, C.K.; Hachiya, T.; Takahara, K.; Kawai-Yamada, M.; Uchimiya, H.; Uesono, Y.; Terashima, I.; Noguchi, K. Effects of AOX1a deficiency on plant growth, gene expression of respiratory components and metabolic profile under low-nitrogen stress in Arabidopsis thaliana. Plant Cell Physiol. 2010, 51, 810–822. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Watanabe, C.K.; Hachiya, T.; Tholen, D.; Shibata, M.; Terashima, I.; Noguchi, K. Distinct responses of the mitochondrial respiratory chain to long- and short-term high-light environments in Arabidopsis thaliana. Plant Cell Environ. 2011, 34, 618–628. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Watanabe, C.K.; Terashima, I.; Noguchi, K. Physiological impact of mitochondrial alternative oxidase on photosynthesis and growth in Arabidopsis thaliana. Plant Cell Environ. 2011, 34, 1890–1899. [Google Scholar] [CrossRef] [PubMed]
- Florez-Sarasa, I.; Flexas, J.; Rasmusson, A.G.; Umbach, A.L.; Siedow, J.N.; Ribas-Carbo, M. In vivo cytochrome and alternative pathway respiration in leaves of Arabidopsis thaliana plants with altered alternative oxidase under different light conditions. Plant Cell Environ. 2011, 34, 1373–1383. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, C.K.A.; Yamori, W.; Takahashi, S.; Terashima, I.; Noguchi, K. Mitochondrial alternative pathway-associated photoprotection of photosystem II is related to the photorespiratory pathway. Plant Cell Physiol. 2016, 57, 1426–1431. [Google Scholar] [CrossRef] [PubMed]
- Dahal, K.; Wang, J.; Martyn, G.D.; Rahimy, F.; Vanlerberghe, G.C. Mitochondrial alternative oxidase maintains respiration and preserves photosynthetic capacity during moderate drought in Nicotiana tabacum. Plant Physiol. 2014, 166, 1560–1574. [Google Scholar] [CrossRef] [PubMed]
- Dahal, K.; Vanlerberghe, G.C. Improved chloroplast energy balance during water deficit enhances plant growth: more crop per drop. J. Exp. Bot. 2018, 69, 1183–1197. [Google Scholar] [CrossRef] [PubMed]
- Giraud, E.; Ho, L.H.M.; Clifton, R.; Carroll, A.; Estavillo, G.; Tan, T.F.; Howell, K.A.; Ivanova, A.; Pogson, B.J.; Millar, A.H.; et al. The absence of ALTERNATIVE OXIDASE1a in Arabidopsis results in acute sensitivity to combined light and drought stress. Plant Physiol. 2008, 147, 595–610. [Google Scholar] [CrossRef]
- Yoshida, K.; Watanabe, C.; Kato, Y.; Sakamoto, W.; Noguchi, K. Influence of chloroplastic photo-oxidative stress on mitochondrial alternative oxidase capacity and respiratory properties: a case study with Arabidopsis yellow variegated 2. Plant Cell Physiol. 2008, 49, 592–603. [Google Scholar] [CrossRef]
- Sakamoto, W.; Miura, E.; Kaji, Y.; Okuno, T.; Nishizono, M.; Ogura, T. Allelic characterization of the leaf-variegated mutation var2 identifies the conserved amino acid residues of FtsH that are important for ATP hydrolysis and proteolysis. Plant Mol. Biol. 2004, 56, 705–716. [Google Scholar] [CrossRef]
- Yoshida, K.; Terashima, I.; Noguchi, K. Up-regulation of mitochondrial alternative oxidase concomitant with chloroplast over-reduction by excess light. Plant Cell Physiol. 2007, 48, 606–614. [Google Scholar] [CrossRef] [PubMed]
- Munekage, Y.; Hojo, M.; Meurer, J.; Endo, T.; Tasaka, M.; Shikanai, T. PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell 2002, 110, 361–371. [Google Scholar] [CrossRef]
- Okegawa, Y.; Kobayashi, Y.; Shikanai, T. Physiological links among alternative electron transport pathways that reduce and oxidize plastoquinone in Arabidopsis. Plant J. 2010, 63, 458–468. [Google Scholar] [CrossRef] [PubMed]
- Takagi, D.; Miyake, C. PROTON GRADIENT REGULATION 5 supports linear electron flow to oxidize photosystem I. Physiol. Plant. 2018, 164, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Shikanai, T. PGR5-dependent cyclic electron flow protects photosystem I under fluctuating light at donor and acceptor sides. Plant Physiol. 2019, 179, 588–600. [Google Scholar] [CrossRef] [PubMed]
- Florez-Sarasa, I.; Noguchi, K.; Araújo, W.L.; Garcia-Nogales, A.; Fernie, A.R.; Flexas, J.; Ribas-Carbo, M. Impaired cyclic electron flow around photosystem I disturbs high-light respiratory metabolism. Plant Physiol. 2016, 172, 2176–2189. [Google Scholar] [CrossRef] [PubMed]
- Oguchi, R.; Ozaki, H.; Hanada, K.; Hikosaka, K. Which plant trait explains the variations in relative growth rate and its response to elevated carbon dioxide concentration among Arabidopsis thaliana ecotypes derived from a variety of habitats? Oecologia 2016, 180, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Funayama, S.; Terashima, I. Effects of geminivirus infection and growth irradiance on the vegetative growth and photosynthetic production of Eupatorium makinoi. New Phytol. 1999, 142, 483–494. [Google Scholar] [CrossRef]
- Fiorani, F.; Umbach, A.L.; Siedow, J.N. The alternative oxidase of plant mitochondria is involved in the acclimation of shoot growth at low temperature. A study of Arabidopsis AOX1a transgenic plants. Plant Physiol. 2005, 139, 1795–1805. [Google Scholar] [CrossRef]
- Kono, M.; Noguchi, K.; Terashima, I. Roles of the cyclic electron flow around PSI (CEF-PSI) and O2-dependent alternative pathways in regulation of the photosynthetic electron flow in short-term fluctuating light in Arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 990–1004. [Google Scholar] [CrossRef]
- Murata, N.; Nishiyama, Y. ATP is a driving force in the repair of photosystem II during photoinhibition. Plant Cell Environ. 2018, 41, 285–299. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.-S.; Liu, M.-J.; Scheibe, R.; Selinski, J.; Zhang, L.-T.; Yang, C.; Meng, X.-L.; Gao, H.-Y. Contribution of the alternative respiratory pathway to PSII photoprotection in C3 and C4 plants. Mol. Plant. 2017, 10, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Shikanai, T.; Yamamoto, H. Contribution of cyclic and pseudo-cyclic electron transport to the formation of proton motive force in chloroplasts. Mol. Plant. 2017, 10, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Hertle, A.P.; Blunder, T.; Wunder, T.; Pesaresi, P.; Pribil, M.; Armbruster, U.; Leister, D. PGRL1 is the elusive ferredoxin-plastoquinone reductase in photosynthetic cyclic electron flow. Mol. Cell 2013, 49, 511–523. [Google Scholar] [CrossRef] [PubMed]
- Dang, K.-V.; Tolleter, J.P.D.; Jokel, M.; Cuiné, S.; Carrier, P.; Auroy, P.; Richaud, P.; Johnson, X.; Alric, J.; Allahverdiyeva, Y.; et al. Combined increases in mitochondrial cooperation and oxygen photoreduction compensate for deficiency in cyclic electron flow in Chlamydomonas reinhardtii. Plant Cell 2014, 26, 3036–3050. [Google Scholar] [CrossRef] [PubMed]
- Bartoli, C.G.; Gomez, F.; Gergoff, G.; Guiamet, J.J.; Puntaru, S. Up-regulation of the mitochondrial alternative oxidase pathway enhances photosynthetic electron transport under drought conditions. J. Exp. Bot. 2005, 56, 1269–1276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, C.K.; Sato, S.; Yanagisawa, S.; Uesono, Y.; Terashima, I.; Noguchi, K. Effects of elevated CO2 on levels of primary metabolites and transcripts of genes encoding respiratory enzymes and their diurnal patterns in Arabidopsis thaliana: possible relationships with respiratory rates. Plant Cell Physiol. 2014, 55, 341–357. [Google Scholar] [CrossRef] [PubMed]
- Dahal, K.; Martyn, G.D.; Alber, N.A.; Vanlerberghe, G.C. Coordinated regulation of photosynthetic and respiratory components is necessary to maintain chloroplast energy balance in varied growth conditions. J. Exp. Bot. 2017, 68, 657–671. [Google Scholar] [CrossRef]
- Florez-Sarasa, I.D.; Bouma, T.J.; Medrano, H.; Azcon-Bieto, J.; Ribas-Carbo, M. Contribution of the cytochrome and alternative pathways to growth respiration and maintenance respiration in Arabidopsis thaliana. Physiol Plant. 2007, 129, 143–151. [Google Scholar] [CrossRef]
- Tohge, T.; Obata, T.; Fernie, A.R. Biosynthesis of the essential respiratory cofactor ubiquinone from phenylalanine in plants. Mol. Plant 2014, 7, 1403–1405. [Google Scholar] [CrossRef]
- Liu, M.; Lu, S. Plastoquinone and ubiquinone in plants: biosynthesis, physiological function and metabolic engineering. Front. Plant Sci. 2016, 7, 1898. [Google Scholar] [CrossRef] [PubMed]
- Genty, B.; Briantais, J.-M.; Baker, N.R. The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta 1989, 990, 87–92. [Google Scholar] [CrossRef]
- Oxborough, K.; Baker, N.R. Resolving chlorophyll a fluorescence images of photosynthetic efficiency into photochemical and non-photochemical components—Calculation of qP and Fv’/Fm’ without measuring Fo’. Photosyn. Res. 1997, 54, 135–142. [Google Scholar] [CrossRef]
- Yoshida, K.; Shibata, M.; Terashima, I.; Noguchi, K. Simultaneous determination of in vivo plastoquinone and ubiquinone redox states by HPLC-based analysis. Plant Cell Physiol. 2010, 51, 836–841. [Google Scholar] [CrossRef] [PubMed]
- Miyagi, A.; Takahashi, H.; Takahara, K.; Hirabayashi, T.; Nishimura, Y.; Tezuka, T.; Kawai-Yamada, M.; Uchimiya, H. Principal component and hierarchical clustering analysis of metabolites in destructive weeds; polygonaceous plants. Metabolomics 2010, 6, 146–155. [Google Scholar] [CrossRef]
- Onda, Y.; Miyagi, A.; Takahara, K.; Uchimiya, H.; Kawai-Yamada, M. Effects of NAD kinase 2 overexpression on primary metabolite profiles in rice leaves under elevated carbon dioxide. Plant Biol. 2014, 16, 819–824. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016; Available online: https://www.R-project.org/ (accessed on 23 June 2019).
| Growth Light Condition | Days after Germination | WT | pgr5 | aox1a/pgr5 | ANOVA p |
|---|---|---|---|---|---|
| HL | 11-day | 0.786 ± 0.041 | 0.772 ± 0.053 | 0.763 ± 0.066 | 0.602 |
| 21-day | 0.791 ± 0.001 * | 0.778 ± 0.007 | 0.770 ± 0.009 | 0.000996 | |
| LL | 15-day | 0.727 ± 0.014 | 0.723 ± 0.013 | 0.647 ± 0.041 * | 0.00324 |
| 30-day | 0.762 ± 0.010 | 0.762 ± 0.007 | 0.761 ± 0.004 | 0.986 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, Z.; Watanabe, C.K.A.; Miyagi, A.; Kawai-Yamada, M.; Terashima, I.; Noguchi, K. Mitochondrial AOX Supports Redox Balance of Photosynthetic Electron Transport, Primary Metabolite Balance, and Growth in Arabidopsis thaliana under High Light. Int. J. Mol. Sci. 2019, 20, 3067. https://doi.org/10.3390/ijms20123067
Jiang Z, Watanabe CKA, Miyagi A, Kawai-Yamada M, Terashima I, Noguchi K. Mitochondrial AOX Supports Redox Balance of Photosynthetic Electron Transport, Primary Metabolite Balance, and Growth in Arabidopsis thaliana under High Light. International Journal of Molecular Sciences. 2019; 20(12):3067. https://doi.org/10.3390/ijms20123067
Chicago/Turabian StyleJiang, Zhenxiang, Chihiro K. A. Watanabe, Atsuko Miyagi, Maki Kawai-Yamada, Ichiro Terashima, and Ko Noguchi. 2019. "Mitochondrial AOX Supports Redox Balance of Photosynthetic Electron Transport, Primary Metabolite Balance, and Growth in Arabidopsis thaliana under High Light" International Journal of Molecular Sciences 20, no. 12: 3067. https://doi.org/10.3390/ijms20123067





